Upcoming workshop on Bayesian Decision Theory and Model Selection at ASA – Alaska Meeting.
July 17th-19th, 2019: A two day workshop discussing Bayesian Decision Theory and Model Selection at the annual meeting of the Alaska Chapter of the American Statistical Association. Co-taught with Drs. Mevin Hooten and Trevor Hefley.
Symposium on integrated population models scheduled at TWS Annual Meeting in 2019.
A ten speaker symposium discussing applications and advances in IPMs scheduled for 1:10-5:00PM on Wednesday, October 2.
Sea otter apex predator paper out in Diversity and Distributions
Williams, PJ, MB Hooten, GG Esslinger, JN Womble, JL Bodkin, MR Bower. 2019. The rise of an apex predator following deglaciation. Diversity and Distributions. 1–14.
Sea Otter Monitoring Protocol for Glacier Bay Complete
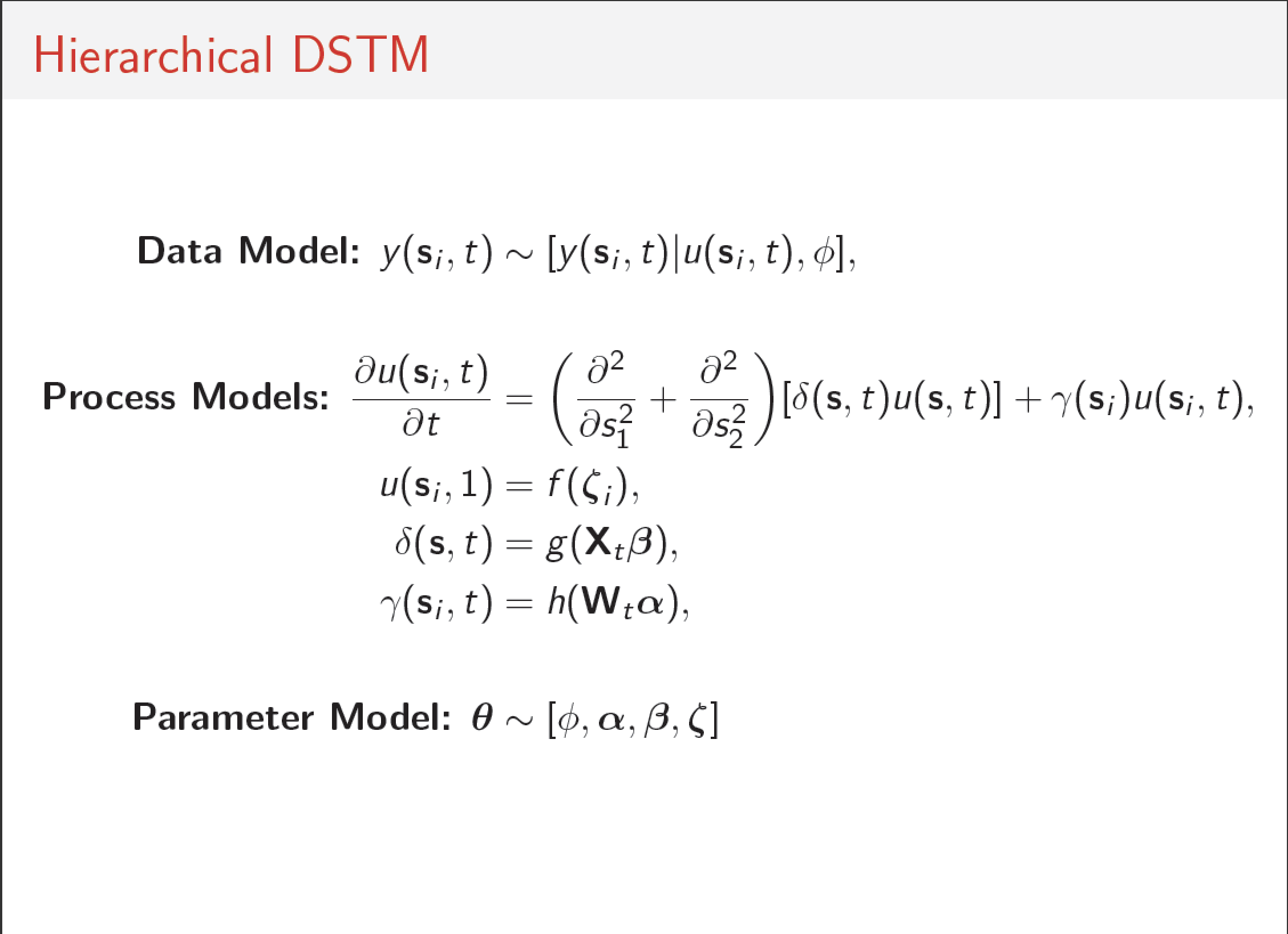
New paper accepted in Ecological Monographs
The paper A guide to Bayesian model checking for ecologists provides a review and description of methods for checking Bayesian hierarchical models.
New paper published in Ecology
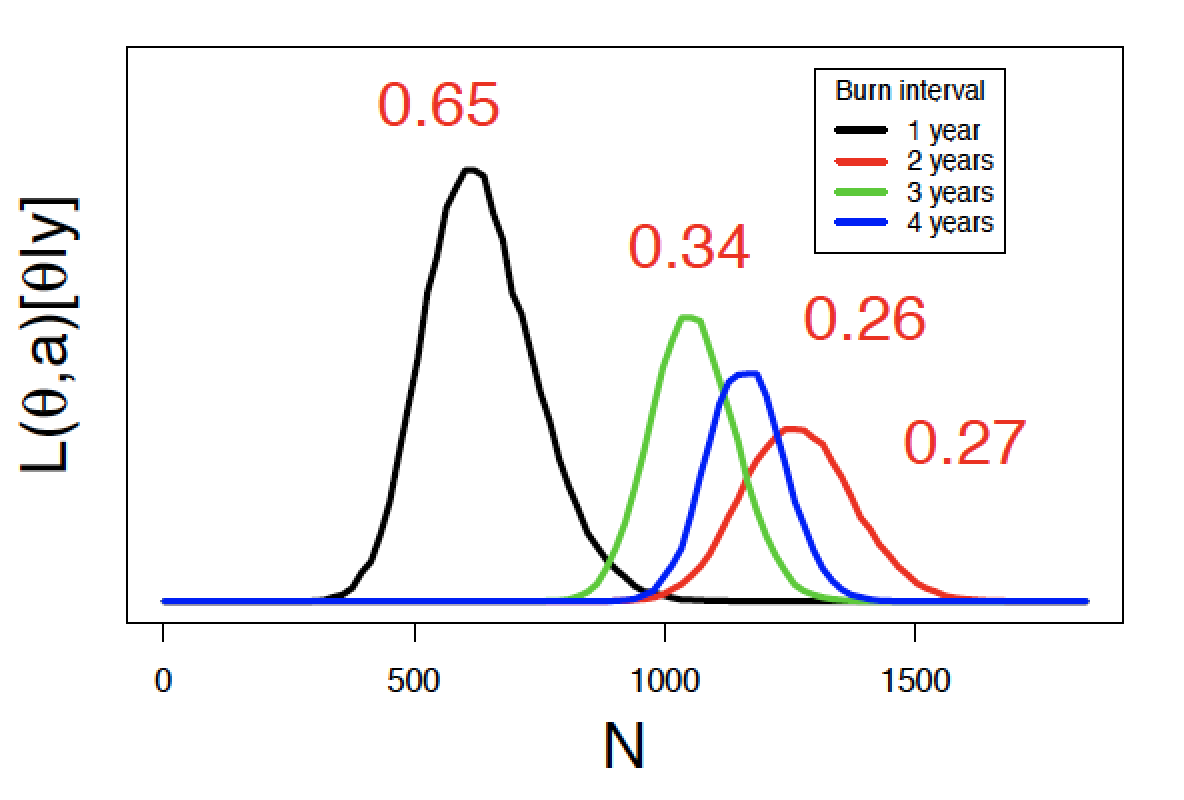
Paper on optimal model-based monitoring of spatio-temporal dynamic processes in early view in Ecology (pdf).
Sea otter image featured on cover of Methods in Ecology and Evolution
New Concepts & Synthesis paper accepted in Ecology on optimal dynamic monitoring
Williams, P. J., M. B. Hooten, J. N. Womble, G. G. Esslinger, and M. R. Bower. In Press. Monitoring dynamic spatio-temporal ecological processes optimally. Ecology (pdf)
Abstract:
Population dynamics varies in space and time. Survey designs that ignore these dynamics may be inefficient and fail to capture essential spatio-temporal variability of a process. Alternatively, dynamic survey designs explicitly incorporate knowledge of ecological processes, the associated uncertainty in those processes, and can be optimized with respect to monitoring objectives. We describe a cohesive framework for monitoring a spreading population that explicitly links animal movement models with survey design and monitoring objectives. We apply the framework to develop an optimal survey design for sea otters in Glacier Bay. Sea otters were first detected in Glacier Bay in 1988 and have since increased in both abundance and distribution; abundance estimates increased from 5 otters to >5,000 otters, and they have spread faster than 2.7 km per year. By explicitly linking animal movement models and survey design, we were able to reduce uncertainty associated with predicted occupancy, abundance, and distribution. The framework we describe is general, and we outline steps to applying it to novel systems and taxa.
A pre-print of “A guide to Bayesian model checking for ecologists” available online
The manuscript is currently in review at Ecological Monographs and a pre-print is available via PeerJ Preprints.
Citation:
(2017) A guide to Bayesian model checking for ecologists. PeerJ Preprints 5:e3390v1https://doi.org/10.7287/peerj.preprints.3390v1
Abstract:
Checking that models adequately represent data is an essential component of applied statistical inference. Ecologists increasingly use hierarchical Bayesian statistical models in their research. The appeal of this modeling paradigm is undeniable, as researchers can build and fit models that embody complex ecological processes while simultaneously controlling observation error. However, ecologists tend to be less focused on checking model assumptions and assessing potential lack-of-fit when applying Bayesian methods than when applying more traditional modes of inference such as maximum likelihood. There are also multiple ways of assessing the fit of Bayesian models, each of which has strengths and weaknesses. For instance, Bayesian p-values are relatively easy to compute, but are well known to be conservative, producing p-values biased toward 0.5. Alternatively, lesser known approaches to model checking, such as prior predictive checks, cross-validation probability integral transforms, and pivot discrepancy measures may produce more accurate characterizations of goodness-of-fit but are not as well known to ecologists. In addition, a suite of visual and targeted diagnostics can be used to examine violations of different model assumptions and lack-of-fit at different levels of the modeling hierarchy, and to check for residual temporal or spatial autocorrelation. In this review, we synthesize existing literature to guide ecologists through the many available options for Bayesian model checking. We illustrate methods and procedures with several ecological case studies, including i) analysis of simulated spatio-temporal count data, (ii) N-mixture models for estimating abundance and detection probability of sea otters from an aircraft, and (iii) hidden Markov modeling to describe attendance patterns of California sea lion mothers on a rookery. We find that commonly used procedures based on posterior predictive p-values detect extreme model inadequacy, but often do not detect more subtle cases of lack of fit. Tests based on cross-validation and pivot discrepancy measures (including the sampled predictive p-value”) appear to be better suited to model checking and to have better overall statistical performance. We conclude that model checking is an essential component of scientific discovery and learning that should accompany most Bayesian analyses presented in the literature.